Scientific Challenge
1| UniZgFall dataset
2| Main goal
3| The scientific challenge
4| Public and private dataset, training and testing examples
5| Input/output formats
6| Performance evaluation
Introduction
Welcome to the IFMBE Scientific Challenge Competition of the 16th Mediterranean Conference on Medical and Biological Engineering and Computing (MEDICON’23). In this edition of the Scientific Challenge participants are asked to distinguish several distinct activities of daily living, based on accelerometer measurements. The motivation includes keeping track of users’ activities in real-time, and possible diagnostics of unwanted and unexpected events, with special attention to movements during human falling and the distinction among various types of falls.
UniZgFall dataset
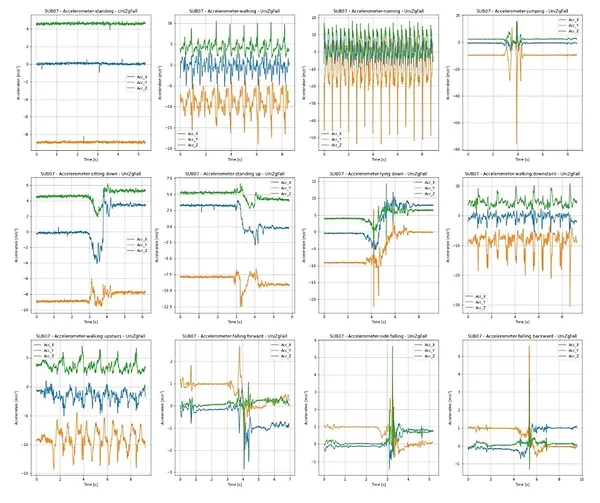
A dataset for accelerometer-based fall detection is used in this Scientific Challenge [1]. For the purpose of data acquisition 16 young healthy subjects were recruited to perform 12 types of activities of daily living (ADL) and 3 types of simulated falls while wearing an inertial sensor unit (Shimmer sensing, Ireland) attached sideways to their waist at belt high. A 2 cm thick tatami mattress was used to cushion the impact during fall simulations. The experiment setting is shown in Figure 1.

Figure 1
Experiment setup for recording the UniZgFall dataset. Subjects wore sensors attached to the belt and performed a set of ADL and simulated falls on a tatami mattress.
[1] D. Razum, G. Seketa, J. Vugrin, and I. Lackovic, “Optimal threshold selection for threshold-based fall detection algorithms with multiple features,” in 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), May 2018, pp. 1513–1516, doi: 10.23919/MIPRO.2018.8400272
The mean and standard deviation for the age, height and weight were 21.9 ± 2.2 years, 178.1 ± 7.8 cm and 70.0 ± 12.1 kg, respectively. The following twelve distinct ADL were performed: walking, fast walking, running, fast running, jumping, high jumping, sitting, standing up, lying down, getting up from lying position, walking down the stairs and walking up the stairs. The three simulated falls were: forward fall, sideways fall and backward fall. Subjects were asked to performed each activity of daily living at least five times (five trials), and for fall activities the subjects performed at least four times. A separate record of acceleration measurements was acquired for each activity trial. The dataset contains 16 folders, one for each subject who participated in the data acquisition process.
An example of accelerometer signals collected during the abovementioned activities of daily living, as well as during the distinct falls, are shown in Figure 2.
Main goal
For the scientific challenge purposes, a set of 10 distinct classes is considered, organized into 5 different groups. The main goal is to developed a classification system, able to classify each record/activity in one of distinct classes {1,2,3,4,5,6,7,8,9,10}.
Group 1 – Moving
- Class 1 | MW – Walking – includes walking and fast walking activities
- Class 2 | MR – Running – includes running and fast running activities
- Class 3 | MJ – Jumping – includes jumping and high jumping activities
Group 2 – Stairs
- Class 4 | WD – Walking down the stairs
- Class 5 | WU – Walking up the stairs
Group 3 – Falls
- Class 6 | FF – Forward fall
- Class 7 | FS – Sideways fall
- Class 8 | FB – Backward fall
Group 4 – Lying down
- Class 9 | LD – Lying down
Group 5 – Inactive
- Class 10 | OT – other classes. This class includes the remaining 3 activities: sitting, standing up and getting up from lying position.
The Scientific Challenge
Quick Start
For those wishing to compete officially, please follow the additional four steps, detailed in i) Rules and Deadlines, ii) Eligibility and iii) Deadlines sections.
- Register using the website of the challenge.
- Download the public data set that you can use to develop your solution and the example source code (MATLAB or Python, respectively challenge.m and challenge.py) to be adapted for your entry.
- Develop your entry by editing the provided files:
-
- Modify the sample entry source code file challenge.m or challenge.py with your changes and improvements.
- Run your modified source code file on all the records in the training and testing data set.
-
- Submit your modified compressed entry file for scoring through the Challenge website. The compressed file has to include all the individual results as described in the expected results.
Improperly-formatted entries will not be scored.
Moreover, we expect the submission of an overview paper with the best solutions to a prestigious Scientific Journal, co-authored by participants of the challenge and considering the publications and discussions at MEDICON’23.
Rules and Deadlines
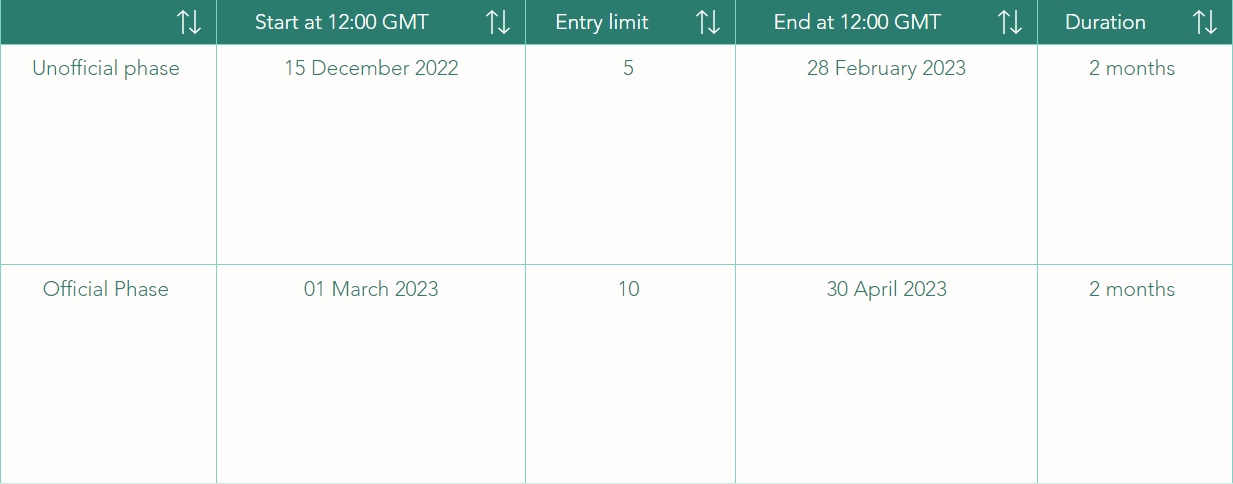
In this challenge, we adopt the general rules of the Physionet Competition. The challenge is structured in two phases: the unofficial (Phase I) and the official (Phase II) phases.
Participants may submit up to 15 entries over both the unofficial and official phases of the competition (see Table 1). Each participant may receive scores for up to five entries submitted during the unofficial phase and ten entries during the official phase. Unused entries may not be carried over to later phases. Entries that cannot be scored (because of missing components, improper formatting, or excessive run time) are not counted against the entry limits.
The unofficial phase (Phase I) will start on 15 December 2022 and will end at 28 February 2023 (2 months). The official phase (Phase II) will start at 01 March 2023 and will end at 30 April 2023 (2 months).
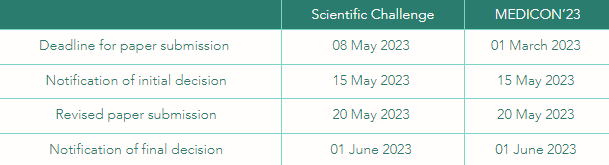
Please note that, to be eligible to the challenge, you should submit a paper to MEDICON’23 describing your approach on the Scientific Challenge to MEDICON’23 (8 May 2023) and, at least, one team member must attend MEDICON’23 and present the paper at the Scientific Challenge Special Session.
Eligibility
All official entries must be received no later than 12:00 GMT on the 30 April 2023. In the interest of fairness to all participants, late entries will not be accepted or scored.
To be eligible you must do all of the following:
1| Register
you should include the username, email address and team name.
2| Submit at least one entry
that can be scored during Phase I until the defined deadline (12:00 GMT, 28 February 2023). All submissions must be in Matlab or Python 3.x and be self-contained (e.g. if some special open-source and non-standard toolbox or library is needed, then it has to be provided. All commercially available toolboxes from Mathworks for MATLAB R2021a are valid). Please follow the detailed information on data formats, input and output formats provided in the sample files.
3| Submit a full paper
describing your work for the Scientific Challenge following the MEDICON’23 rules no later than 08 May 2023 using the regular submission system. Include the overall score for at least one Phase I or Phase II entry in your paper. You will be notified if your paper has been accepted by email from MEDICON’23 by 15 May 2023 (initial decision) and by 01 June 2023 (final decision).
4| Attend MEDICON’23 (September 14-16, 2023)
and present your work there (at least one team member must attend MEDICON’23).
Please do not submit analysis of this year’s Scientific Challenge data to other conferences or journals until after MEDICON’23 has taken place, so the competitors are able to discuss the results in a single forum. We expect the submission of an overview paper with the best solutions to a prestigious Scientific Journal, co-authored by participants of the challenge and taking into account the publications and discussions at MEDICON’23.
Deadlines
Scientific challenge – Phases
Scientific Challenge / MEDICON’23 – alignment
Public and private dataset, training and testing examples
- Public and private datasets
A public dataset will be available in the unofficial phase, containing 3 training examples. Additional training examples may be made available in the official phase.
In parallel, a private dataset (testing set) will be kept by the organization team to compute the participant’s scoring purposes.
- Training and testing examples
Training examples
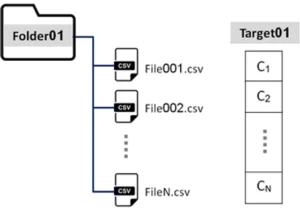
Folder and records: For the scientific challenge purpose, a training example consists of a folder, containing a variable number (N) of records/files, each one corresponding to a distinct activity performed by the same participant. Each record/file (CSV – comma-separated values format) is composed of N rows and 4 columns: the first three columns correspond to the X, Y, and Z accelerometer values; the last is the time stamp.
File format
- Target: Moreover, for each folder a target vector is provided, with the same dimension of the number of records (N), where each value identifies the type of activity performed by the participants (class).
Target={C_1,C_2,…,C_N }
with C_i={1,2,3,4,5,6,7,8,9,10}
Figure 3 illustrates one training example: Folder and respective files; target C_i.
- Testing examples
The testing examples are similar to the training examples. They consist of a folder containing a variable number of files, each one corresponding to a specific activity.
However, note that in a testing example the activities (files) can belongs to different participants. Moreover, in opposition to the training examples, the target vector is not provided in the testing examples.
Input/output formats
- Inputs
To enter the MEDICON’23 challenge, the participants have to develop an algorithm that is able to read all files N in a folder (time and XYZ values), and provide an output vector of dimension N: the respective class for each activity/file. The algorithm must be implemented in MATLAB or Python 3.x.
For clarification purposes, an example can be download from the MEDICON’23 challenge website (example. zip archive) containing:
- Folder: consists of a folder containing N csv files, each one composed of 4 columns: {X, Y, Z, time}
- Target.txt: the target vector (txt file), with dimension N, where each value identifies the specific activity performed by the participant, classified as {1,2,3,4,5,6,7,8,9,10}.
- challenge.py: example of a Python source code, that uses the folder (and files inside the folder) to compute the result vector. This result vector is compared with the target vector to obtain the final score.
- challenge.m: example of a Matlab source code, that uses the folder (and files inside the folder) to compute the result vector. This result vector is compared with the target vector to obtain the final score.
- Result.txt: vector file, containing the result obtained with the Matlab/Python program.
■ Outputs – Entry to be submitted to the challenge
Each team must upload to the MEDICON’23 challenge website a .zip, .7z, .rar or tar.gz archive containing the following files:
1| AUTHORS.txt: a plain text file listing the members of your team who contributed to your code, and their affiliations.
2| Code: All the required code that is used for the analysis. This set of files must include a file with the name challenge.m or challenge.py, which is the main entry file of the code and accepts one input: a record name.
3| Result: Your code should output the file resultFolderNumber.txt, where FolderNumber is the name of the Folder data set that was used for the analysis. As described, the result file should consist of a text file, one value per line, where each value identified the obtained class {1,2,3,4,5,6,7,8,9,10} for each activity.
We will run your code on the testing set and compare the contents of the resultFolderNumber.txt files that are generated with the ground-truth annotations (targetFolderNumber.txt). If everything runs as expected, you will get an email with the score of your submission.
In Phase I (Unofficial) your submissions will be tested on a subset of the testing set, while in Phase II (Official) they will be tested on the whole testing set. Thus, the grade for a specific algorithm may be different in each phase.
Performance evaluation
The overall score for each entry (a Folder) is computed based on the resultFolderNumber.txt file you provided.
In the Scientific Challenge we use F1score metric as basis:
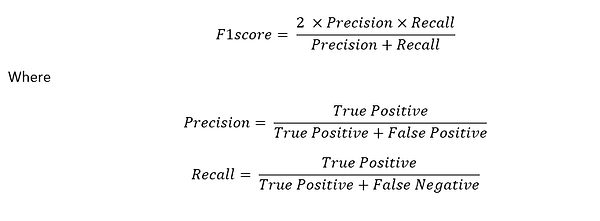
Score example
Let us assume a folder (Folder01) containing N=30 records/files, each one corresponding to a specific activity. The target vector (targetFolder01.txt) is defined as:
Target = [1,1,3,4,10,10,3,6,7,8,7,7,7,7,8,10,10,10,10,10,10,1,1,2,2,3,3,3,3,3]
Let us assume a result vector (resultFolder01.txt) defined as:
Result = [1,1,2,2,10,3,5,6,7,8,7,7,8,8,8,10,10,10,10,1,1,1,1,2,6,6,6,6,3,3]
From the target and result vectors it is possible to build a confusion matrix:
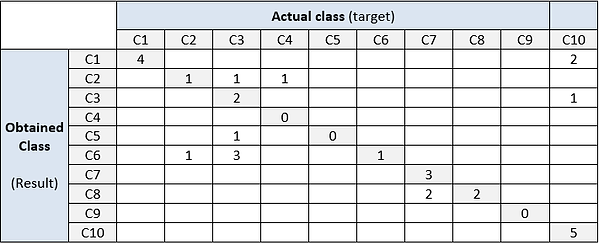
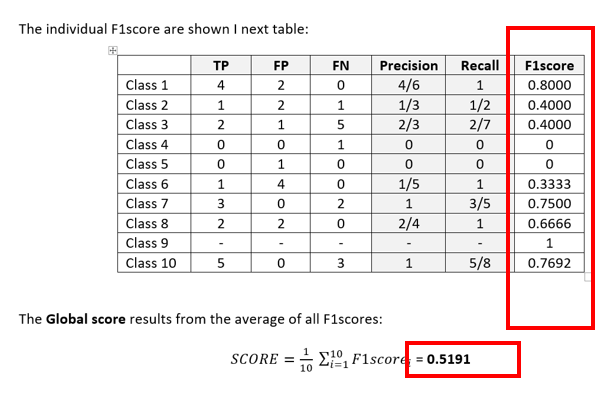
Award
The scientific challenge will award a total of €1.000 (one thousand Euros) in scientific prizes.
Contact us Email: [email protected]